Enhancing Exon Skipping Efficiency: A Predictive Computational Approach
Duchenne muscular dystrophy (DMD) is a severe genetic disorder caused by mutations in the DMD gene, leading to loss of dystrophin protein and progressive muscle degeneration. One promising therapeutic approach is exon skipping, which uses antisense oligonucleotides (AONs) to restore the reading frame of dystrophin transcripts. However, selecting optimal AON sequences remains a challenge.
A Novel Computational Approach
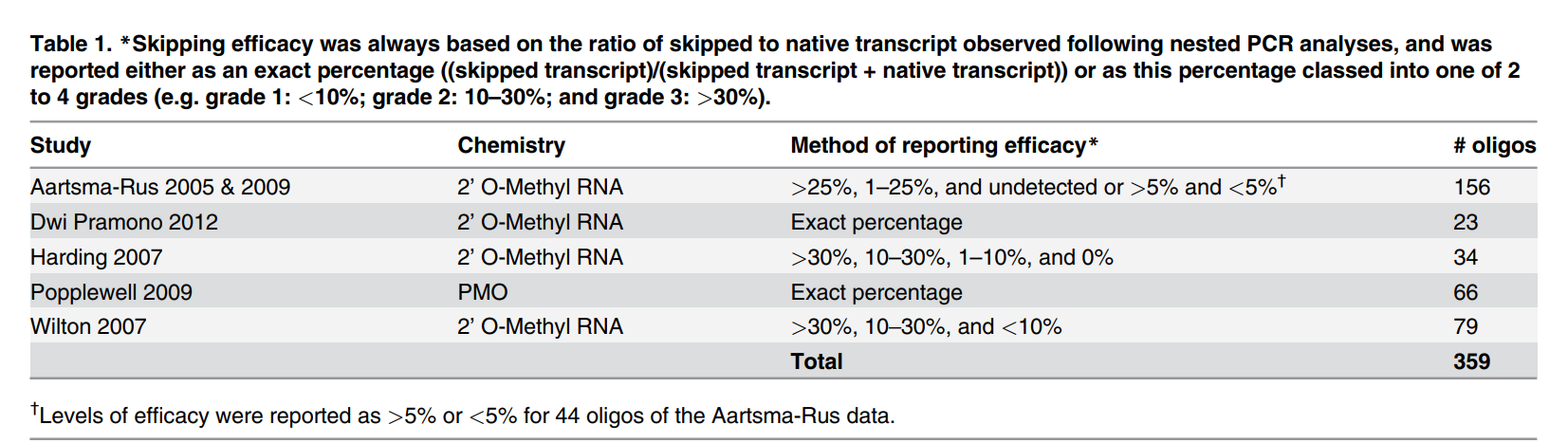
This study presents a novel in silico pre-screening approach that applies predictive statistical modeling to evaluate exon skipping efficiency. By compiling a large dataset of previously tested oligonucleotides, researchers identified two key parameters influencing exon skipping:
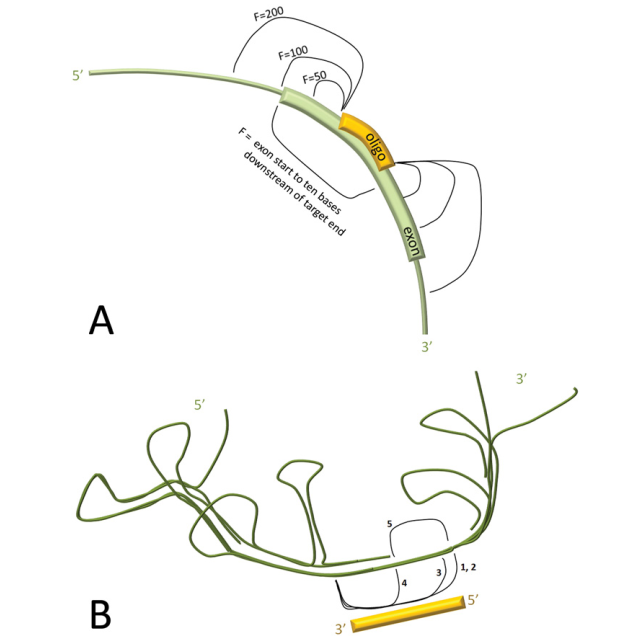
- Binding energetics of the oligonucleotide to the target RNA
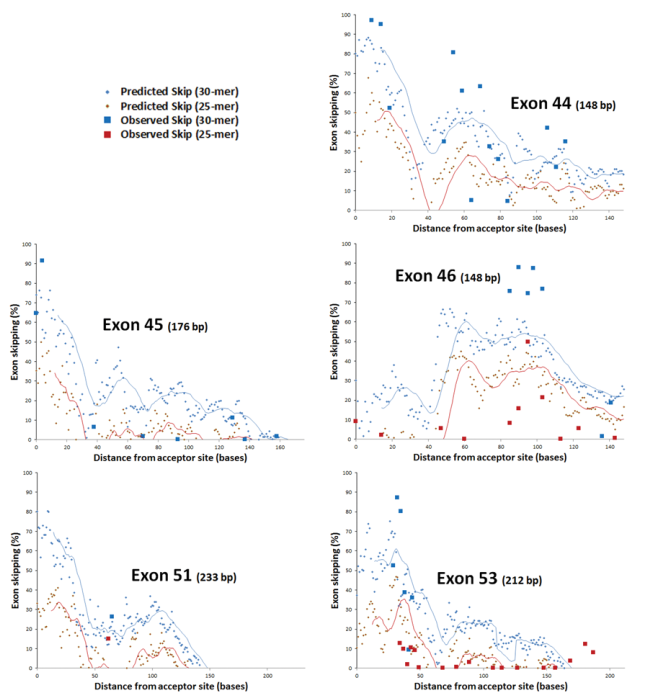
- Distance of the target site from the splice acceptor site
Algorithm Performance and Validation
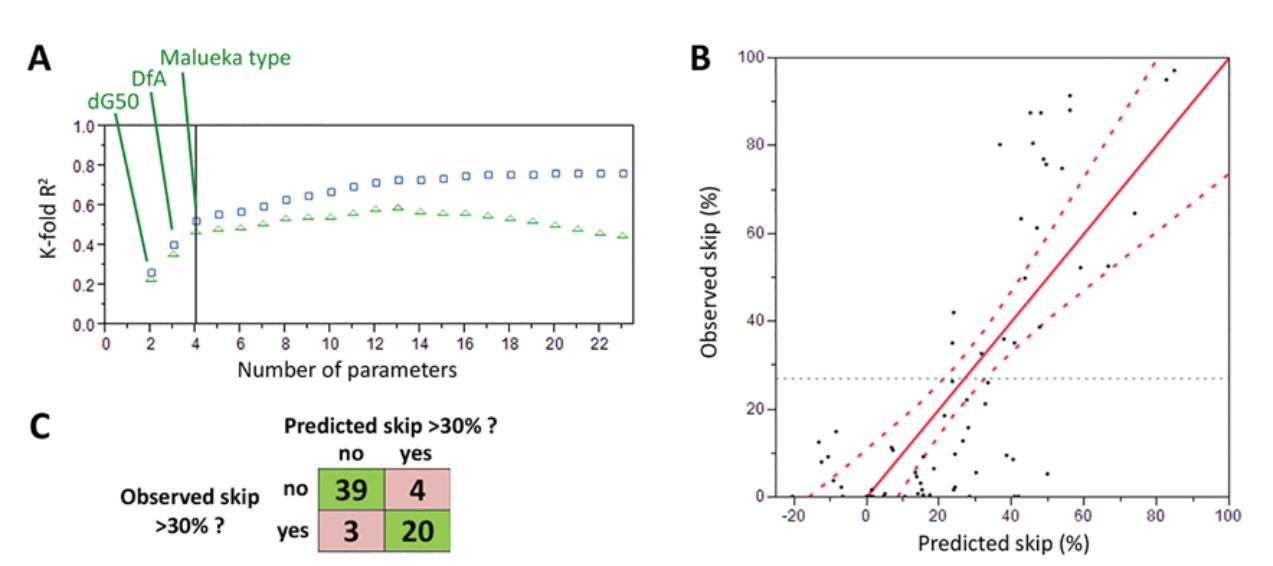
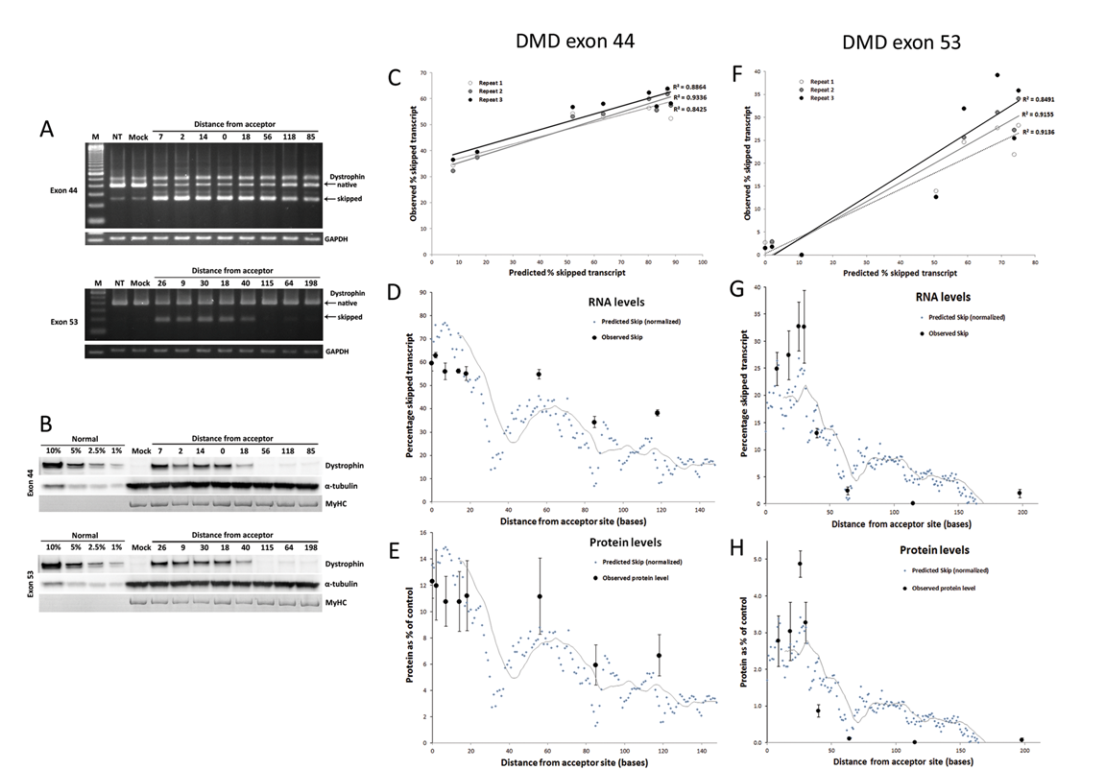
Using these insights, the team developed predictive algorithms that correctly classified high- and low-efficacy sequences with 89% accuracy for phosphorodiamidate morpholino oligomers (PMOs) and 76% accuracy for 2’-O-Methyl RNA oligonucleotides. These predictions were validated through in vitro exon skipping assays, which demonstrated strong correlation with experimental results (R² = 0.89).
Key Findings
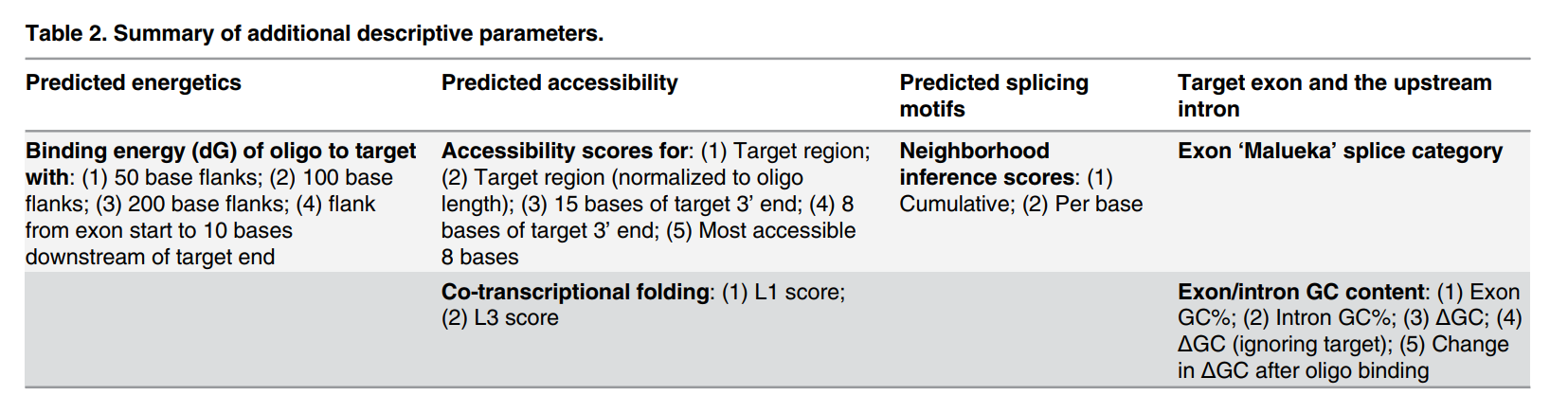
- Exon skipping efficacy correlates with binding free energy (ΔG) and target site positioning.
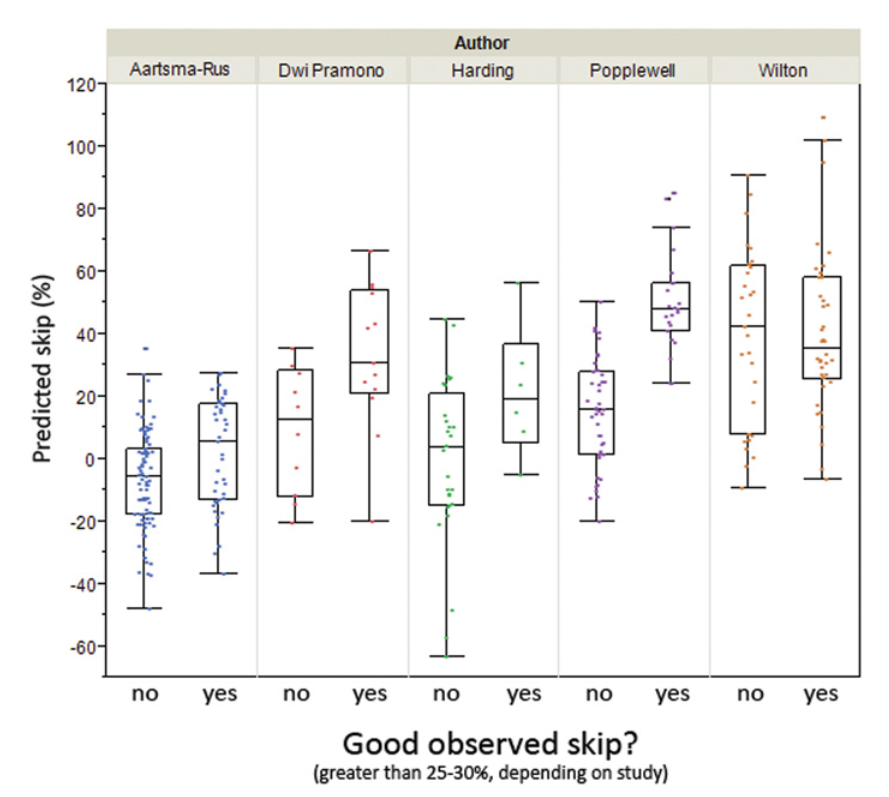
- The Malueka exon category affects skipping efficiency for PMOs but not for 2'-O-Methyl oligonucleotides.
- An in silico screening tool was developed to predict exon skipping across any target exon.
Conclusion
This study provides a computational framework for rational oligonucleotide design, improving efficiency in therapeutic exon skipping strategies for DMD and potentially other genetic disorders.





| Field | Details |
| Title | In Silico Screening Based on Predictive Algorithms as a Design Tool for Exon Skipping Oligonucleotides in Duchenne Muscular Dystrophy |
| Authors | Yusuke Echigoya, Vincent Mouly, Luis Garcia, Toshifumi Yokota, William Duddy |
| Corresponding Author | William Duddy, Toshifumi Yokota |
| Publication Date | 27-Mar-15 |
| Journal | PLOS ONE |
| Keywords | Exon skipping, Duchenne muscular dystrophy, antisense oligonucleotides, in silico screening, predictive algorithms |
| Methods Used | Predictive statistical modeling, computational analysis, in vitro validation, RT-PCR, Western blot |
| Study Type | Research article |
| DOI | 10.1371/journal.pone.0120058 |